Survival analysis¶
A popular technique to perform a survival analysis (regression) is the Cox’s model. It is possible to perform such an analysis using imputation data (dosage format), where each imputed genotypes varies between 0 and 2 (inclusively). A value close to 0 means that a homozygous genotype of the most frequent allele is the most probable. A value close to 2 means that a homozygous genotype of the rare allele is the most probable. Finally, a value close to 1 means that a heterozygous genotype is the most probable.
We suppose that you have followed the main Genome-wide imputation pipeline. The following command will create the working directory for this tutorial.
mkdir -p $HOME/genipe_tutorial/survival
Input files¶
Imputed genotypes¶
After running the genipe pipeline, the imputed genotypes files will
have the .impute2 or .impute2.gz extension Those files will be located
in the final_impute2 directories of each chromosomes. There should be one
impute2 file per chromosome (see the
genipe/chrN/final_impute2 directories section in the main
Genome-wide imputation pipeline). These files consist of the imputed genotypes required
to perform the analysis.
The general structure of the file contains the following columns (which are space delimited): the chromosome, the name of the marker, its position and its two alleles. The subsequent columns correspond to the probabilities of each genotype (hence, there are three columns per sample). The first value correspond to the probability of being homozygous of the first allele. The second value correspond to the probability of being heterozygous. Finally, the third value correspond to the probability of being homozygous of the second allele. The following example shows two lines of the impute2 file.
21 rs376366718:10000302:A:G 10000302 A G 0.986 0.014 0 1 0 0 1 0 0 ...
21 21:10002805:C:T 10002805 C T 0.254 0.736 0.010 0.810 0.188 0.002 0.800 0.195 0.005 ...
Samples file¶
This file is generated by genipe and has the .sample extension.
There should be one sample file per chromosome (see the
genipe/chrN/final_impute2 directories section in the main
Genome-wide imputation pipeline). These files greatly resembles the Plink fam
file. Specifically, it contains the samples that are included in the impute2
file (with the same order). It is needed to correctly interpret the sample
described by the impute2 file. The format is as follow:
ID_1 ID_2 missing father mother sex plink_pheno
0 0 0 D D D B
1341 NA06985 0 0 0 2 -9
1341 NA06991 0 NA06993 NA06985 2 -9
1341 NA06993 0 0 0 1 -9
...
The first two rows are part of the format and should be as is.
Warning
The column ID_2 should contain unique sample identification numbers,
since the analysis will only consider the ID_2 (which correspond to the
sample ID in the Plink file) to correctly match the samples and the
imputed genotypes.
Phenotype file¶
This file describes the variables used to perform the survival regression. The
file is tab separated and contains one row per sample, one column per
variable. Two variables are required: the time to event, and the event (1,
the event was observed, and 0, the event was not observed
(censored)).
The following is an example of a phenotype file:
SampleID TTE Event Age Var1 Gender
NA06985 425.5006566841411 1 53 48.01043142060001 2
NA06993 553.6637748799277 1 47 23.7615117523 1
NA06994 569.4273004275149 0 48 20.2946857226 1
We provide a dummy phenotype file (where values, except for Gender, were
randomly generated). The following command should download the phenotype file.
cd $HOME/genipe_tutorial/survival
wget http://pgxcentre.github.io/genipe/_static/tutorial/phenotypes_survival.txt.bz2
bunzip2 phenotypes_survival.txt.bz2
Note
Note that the gender is encoded such that males are 1 and females are
2. Samples with missing gender (encoded as 0) will be excluded only
if gender is in the covariable list.
Warning
The sample identification numbers should match the ones in the sample file (see above). Those numbers should be unique for each sample. Only the samples that are both in the sample and phenotype files will be kept for analysis. The order of the samples in the phenotype file is not important.
Sites to extract (optional)¶
This file (which is optional) should contain a list of site (one identification
number per line) to keep for the analysis. This file might be the
.good_sites file automatically generated by genipe (see the
genipe/chrN/final_impute2 directories section in the main
Genome-wide imputation pipeline).
Executing the analysis¶
If you followed the Genome-wide imputation pipeline, the following commands should execute the survival analysis.
cd $HOME/genipe_tutorial/survival
imputed-stats cox \
--impute2 ../genipe/chr22/final_impute2/chr22.imputed.impute2.gz \
--sample ../genipe/chr22/final_impute2/chr22.imputed.sample \
--pheno phenotypes_survival.txt \
--extract-sites ../genipe/chr22/final_impute2/chr22.imputed.good_sites \
--nb-process 8 \
--nb-lines 6000 \
--gender-column Gender \
--covar Age,Var1,Gender \
--sample-column SampleID \
--time-to-event TTE \
--event Event
For more information about the arguments and options, see the Usage section. For an approximation of the execution time, refer to the Statistical Analysis Execution Time section.
Output files¶
There will be two output files: .cox.dosage will contain the statistics,
and .log will contain the execution log.
.cox.dosage file¶
This file contains the results from the survival analysis. It shows the following information:
chr: the chromosome.pos: the position on the chromosome.snp: the name of the marker.major: the major allele.minor: the minor allele.maf: the frequency of the minor allele.n: the number of samples that were used for this marker.coef: the coefficient.se: the standard error.lower: the lower value of the 95% confidence interval.upper: the upper value of the 95% confidence interval.z: the z-statistic.p: the p-value.
Note
By default, the statistics are computed only for markers with a minor allele
frequency of 1% and higher. Markers with lower MAF will have NA values.
To modify this behavior, use the --maf option.
Usage¶
The following command will display the documentation for the survival analysis in the console:
$ imputed-stats cox --help
usage: imputed-stats cox [-h] [-v] [--debug] --impute2 FILE --sample FILE
--pheno FILE [--extract-sites FILE] [--out FILE]
[--nb-process INT] [--nb-lines INT] [--chrx]
[--gender-column NAME] [--scale INT] [--prob FLOAT]
[--maf FLOAT] [--covar NAME] [--categorical NAME]
[--missing-value NAME] [--sample-column NAME]
[--interaction NAME] --time-to-event NAME --event
NAME
Performs a survival regression on imputed data using Cox's proportional hazard
model. This script is part of the 'genipe' package, version 1.4.2.
optional arguments:
-h, --help show this help message and exit
-v, --version show program's version number and exit
--debug set the logging level to debug
Input Files:
--impute2 FILE The output from IMPUTE2.
--sample FILE The sample file (the order should be the same as in
the IMPUTE2 files).
--pheno FILE The file containing phenotypes and co variables.
--extract-sites FILE A list of sites to extract for analysis (optional).
Output Options:
--out FILE The prefix for the output files. [imputed_stats]
General Options:
--nb-process INT The number of process to use. [1]
--nb-lines INT The number of line to read at a time. [1000]
--chrx The analysis is performed for the non pseudo-autosomal
region of the chromosome X (male dosage will be
divided by 2 to get values [0, 0.5] instead of [0, 1])
(males are coded as 1 and option '--gender-column'
should be used).
--gender-column NAME The name of the gender column (use to exclude samples
with unknown gender (i.e. not 1, male, or 2, female).
If gender not available, use 'None'. [Gender]
Dosage Options:
--scale INT Scale dosage so that values are in [0, n] (possible
values are 1 (no scaling) or 2). [2]
--prob FLOAT The minimal probability for which a genotype should be
considered. [>=0.9]
--maf FLOAT Minor allele frequency threshold for which marker will
be skipped. [<0.01]
Phenotype Options:
--covar NAME The co variable names (in the phenotype file),
separated by coma.
--categorical NAME The name of the variables that are categorical (note
that the gender is always categorical). The variables
are separated by coma.
--missing-value NAME The missing value in the phenotype file.
--sample-column NAME The name of the sample ID column (in the phenotype
file). [sample_id]
--interaction NAME Add an interaction between the genotype and this
variable.
Cox's Proportional Hazard Model Options:
--time-to-event NAME The time to event variable (in the pheno file).
--event NAME The event variable (1 if observed, 0 if not observed)
Results comparison¶
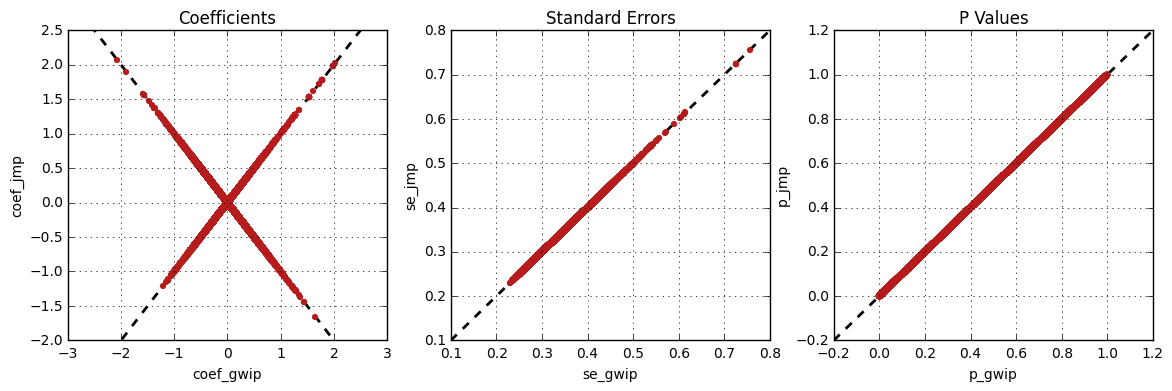
The survival analysis results from genipe and JMP Genomics were
compared for validity. The following figure shows the comparison for, from left
to right, the coefficients, the standard errors and the p-values. The x
axis shows the results from genipe, and the y axis shows the
results for JMP Genomics. This comparison includes 48,127 “good” imputed
markers with a MAF higher or equal to 15%, analyzed for 60 samples (i.e
results from this tutorial). Note that for this comparison, the probability
threshold (--prob) was changed from 0.9 to 0 to imitate JMP
Genomics analysis (see note below for more information).
Note
Only markers with minor allele frequency (MAF) higher or equal to 15% were compared, since markers with lower MAF might have convergence issues (e.g. all exposed samples are all cases or all controls). In that case, the coefficient is large.
Note
The sign of the coefficients might be different when comparing
genipe to GMP Genomics, since genipe computes the
statistics on the rare allele, while JMP Genomics computes them on the
second (alternative) allele. The alternative allele might not always be the
rarest.
Note
By default, genipe excludes samples with a maximum probability
lower than 0.9 (the --prob option), while JMP Genomics keeps all the
samples for the analysis. In order to get the same results as JMP
Genomics, the analysis must be done with a probability threshold of 0
(i.e. --prob 0, keeping all imputed genotypes including those with
poor quality). This is what was done for the previous figure.
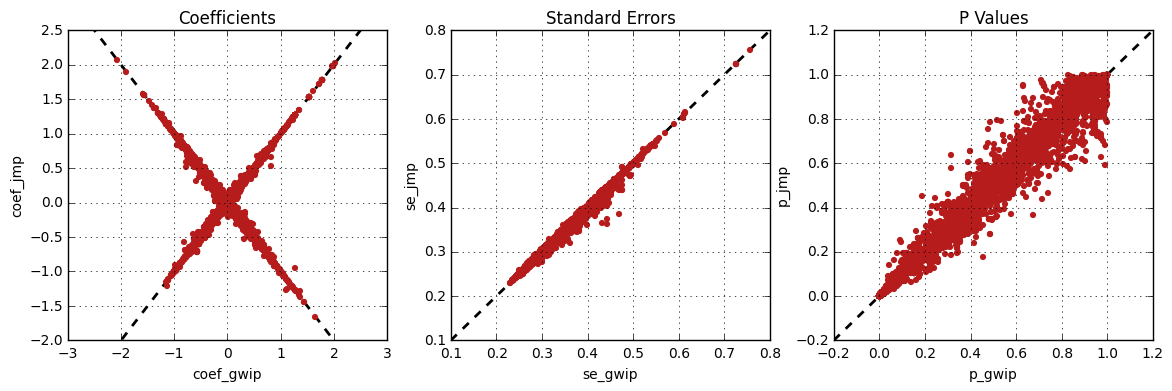
The following figure shows the comparison between JMP Genomics and
genipe for the same analysis, but using the default probability
threshold of 0.9 (excluding imputed genotypes with poor quality). Hence,
48,045 markers were compared.