Statistical Analysis Execution Time¶
GWAS analysis of imputed markers is computationally intensive. While it is feasible to run such analyses on some simple models like linear and logistic regression, more complex models like Cox regression and mixed linear models require more computing power or specialized implementations.
We have optimized the mixed linear model analysis to significantly decrease computation time. Using a two-step approach (as described by Sikorska et al., 2015 [doi: 10.1038/ejhg.2015.1]), the execution time is comparable to a simple linear regression. Prior to optimization, the analysis of chromosome 2 was performed in 53 hours for 33 sub-analysis with 6 threads each (which corresponds to 198 threads).
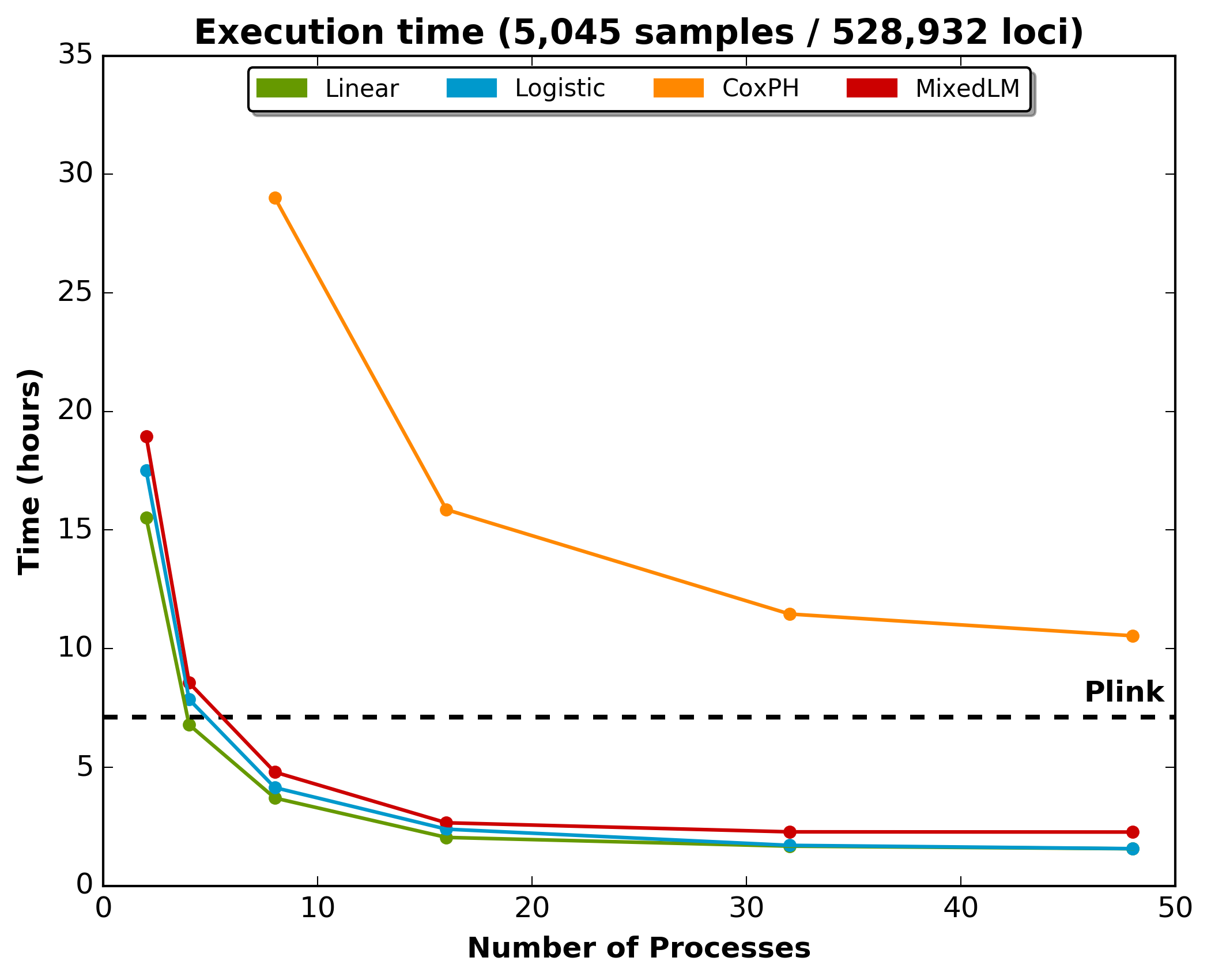
The following figure shows the execution time for a typical imputation analysis of chromosome 2, imputed for 5,045 samples. Chromosome 2 was composed a total of 1,170,797 loci, where 961,019 were of sufficient quality, and 528,932 had a MAF higher than 1%. The black dashed line is the execution time for Plink.
Note
Depending on the installation, when executing the analysis with n threads, OPENBLAS (typical python environment) or MKL (miniconda environment) automatically use all the CPUs for each thread, such that the load quickly increases to n times the number of CPUs. Such high load slows down the analysis considerably.
To avoid this, always export the following environment variables and specify
the total number of threads using the --nb-process option.
export OPENBLAS_NUM_THREADS=1
export MKL_NUM_THREADS=1
We are planning to optimize the Cox’s proportional hazard regression in the near future.